Abstract
Introduction: Next-generation sequencing (NGS) has led to the generation of massive amounts of sequencing data from cancer samples, including MPNs and MDS/MPNs. Current publications a vast heterogeneity among tumor genotypes across different patients. However, analysis of patterns of mutations correlation can reveal some genes that are more likely to co-occur in the same patients, while other genes tend to be mutually exclusive and are almost never found in the same case. Analysis of patterns of co-occurrence and mutual exclusivity of gene mutations can lead to insights in the pathogenesis of these diseases and can help identify putative novel driver genes that are mutually exclusive from known driver genes.
Patients and Methods:
We analyzed data of 646 patients with MPN or MDS/MPN, including 210 patients from our center and 436 patients with data that was available in the literature. Patients from our center were analyzed through either WES (N=123) or targeted sequencing panel of 253 genes involved in myeloid malignancies (N=87). Among the 436 cases selected from published studies in the literature, 278 had been analyzed by WES and 158 were evaluated by large sequencing panels containing the majority of myeloid related genes. We sought to analyze a common denominator of 26 genes that found to be recurrently patients: JAK2, TET2, ASXL1, SRSF2, CALR, DNMT3A, SF3B1, CBL, NRAS, KRAS, PTPN11, EZH2, U2AF1, RUNX1, CUX1, MPL, ZRSR2, TP53, IDH2, SH2B3,PHF6, GATA2, IDH1, SETBP1, ETV6, STAG2 . For the 210 patients included from our center, reads were aligned and processed following Genome Analysis Toolkit best practices, and somatic variants were called combining the output of SomaticSniper, Mutect and Pindel for WES data and Mutect/Mutect2 for targeted panel data. Regarding public data, we selected 7 studies that evaluated patients with MPNs and MDS/MPNs through WES/targeted sequencing and where sample-level data on all individual mutations that were identified were available. Pairwise analysis of occurrence and mutual exclusivity used Fisher tests with p-values adjusted for false discovery rate using the Benjamini-Hochberg method with a q-value cutoof of 0.1, while Multi-Dendrix (Raphael Lab, Brown University) was used for determination of groups of mutual exclusive genes.
Results:
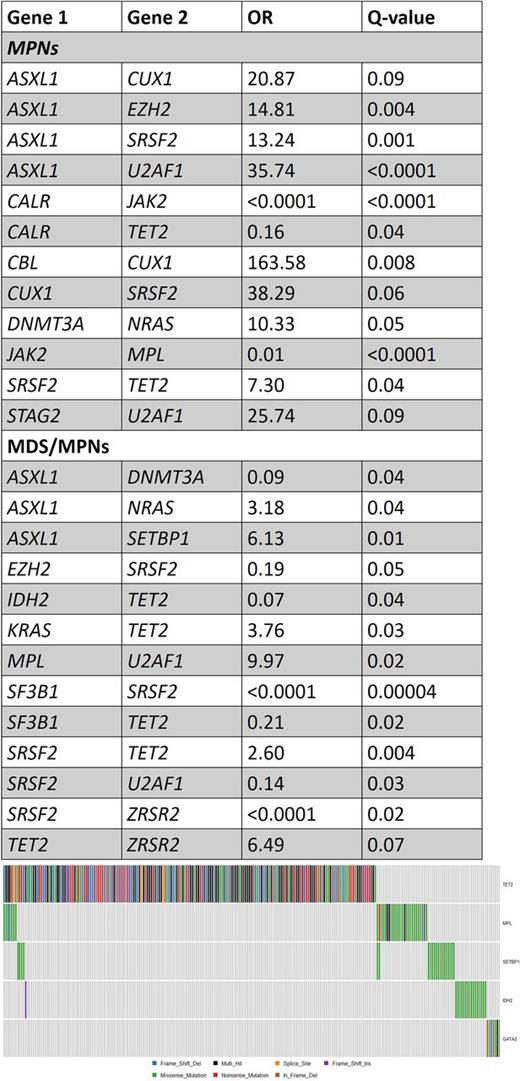
Diagnosis of the cohort included essential thrombocythemia (ET; N=134), polycythemia vera (PV; N=113), myelofibrosis (MF; N=123), chronic myelomonocytic leukemia (CMML; N=228), atypical CML (aCML; N=15) and more rare disorders (MPN-unclassified [N=4], MDS/MPN-unclassified [N=10], refractory anemia with ring sideroblasts and thrombocytosis [N=18] and chronic neutrophilic leukemia [N=1]). Since some genes tend to co-occur more frequently with a specific disease type (e.g. JAK2 in Polycythemia Vera) we analyzed cases of MPNs and MDS/MPNs separately. Results are summarized in the Table. Among patients with MPNs, some novel associations appear: ASXL1 tends to co-occur with CUX1, EZH2, SRSF2 and U2AF1, while CALR and TET2 mutations tend to be mutually exclusive. SRSF2 mutations tend to co-occur with TET2 and CUX1 mutations, and U2AF1 with STAG2 mutations. In patients with MDS/MPNs, different associations are seen, including a tendency for mutual exclusivity of ASXL1 and DNMT3A mutations (Odds Ratio [OR]=0.09) and TET2 and SF3B1 mutations (OR=0.21), while TET2 t ends to co-occur with KRAS, SRSF2 and ZRSR2, while MPL associates strongly with U2AF1 mutations (OR=9.97). Analysis of sets of mutual exclusive genes with Multi-Dendrix revealed the following sets of genes that tend to be mutually exclusive (considering the whole cohort): Set 1- CALR, CBL, CUX1, JAK2, NRAS, RUNX1; Set 2 - GATA2, IDH2, MPL, SETBP1, TET2; Set 3- EZH2, SF3B1, SRSF2, TP53, U2AF1, ZRSR2; Set 4- ASXL1, DNMT3A, IDH1, KRAS, PHF6. Set 2 is illustrated in Figure 1.
Conclusions:
Despite a vast array of different mutations and possible combinations in MPNs and MDS/MPNs, it is clear that some combinations of gene mutations occur more frequently than others, while some genes almost never co-occur together. Analysis of the functional consequences of frequent gene associations might lead to improvements in diagnostic and molecular classification of these neoplasms. Larger cohorts, in the range of thousands of patients, are needed to better determine the more prevalent patterns of co-association and mutual exclusivity.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.